Sensitivity meets speed
Rapidly prepare stranded, whole transcriptome sequencing libraries using Watchmaker RNA Library Prep Kits with Polaris™ Depletion. The highly streamlined Polaris Depletion module depletes highly abundant rRNA and globin transcripts in human, mouse, and rat samples, providing improved coverage of biologically interesting transcripts, including long non-coding RNAs.
The automation-friendly workflow reduces total turnaround time, hands-on time, and consumable use through a reduction in bead purification and reaction steps. A novel, engineered reverse transcriptase improves the conversion of RNA to cDNA, enabling high-quality performance with 1 ng to 1000 ng of total RNA, as well as FFPE-derived RNA.
Key Features and Benefits
- Increase sensitivity with clinically relevant samples, including FFPE and inputs as low as 1 ng
- Generate high-complexity libraries in under 4.5 hours, including rRNA and globin depletion
- Improve automatability with fewer purifications and reagent additions
- Consolidate disparate/niche workflows into one process with broad sample type and input compatibility
- Realize the performance benefits of a reverse transcriptase specifically tailored for RNA-seq applications
Watch now: Interrogating Tumor Structural Variation and Nongenic Dark Matter with Whole-Transcriptome Profiling
As presented on September 17, 2024
Applications
- RNA sequencing with degraded samples, such as FFPE
- Detection of long non-coding RNAs (lncRNAs)
- Gene fusion, splice variant and isoform detection
- Gene expression analysis
- Novel transcript discovery
- Single nucleotide variant (SNV) detection
- Targeted sequencing protocols employing hybridization capture
Process more samples easily with a simple, rapid workflow
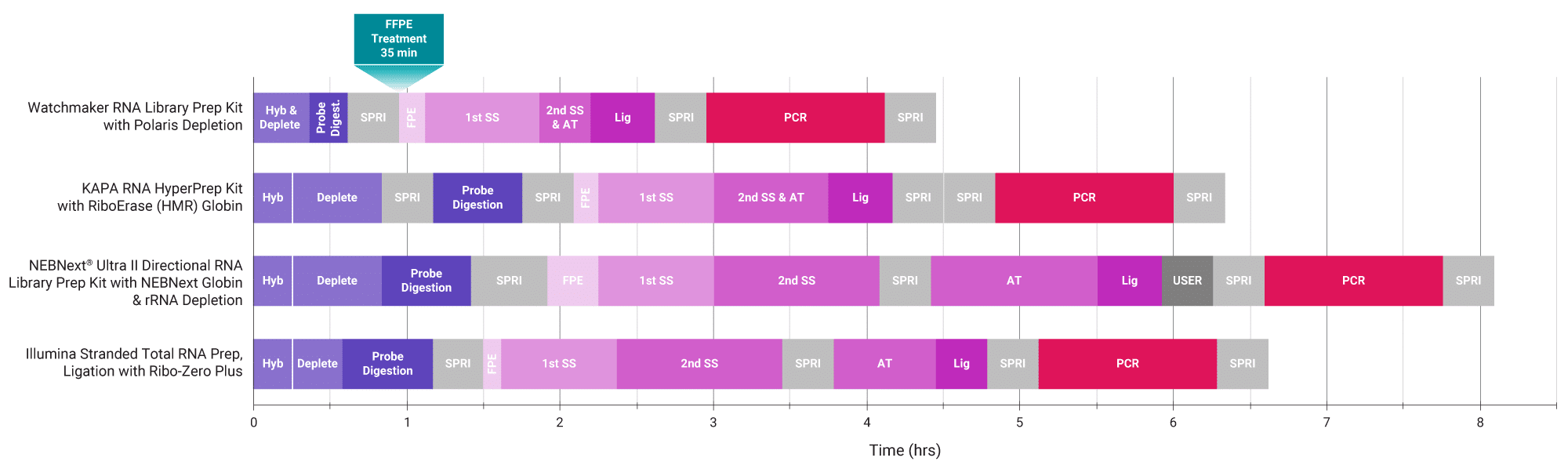
Figure 1. Improve automatability and reduce turnaround time. The Watchmaker solution combines and shortens enzymatic steps and has fewer bead purifications in comparison to commercially-available kits, resulting in a highly automatable workflow with significantly reduced hands-on time (up to one hour per plate) and consumable use (up to 1,000 pipette tips per plate).
Key Performance Data
Access more information from FFPE samples
Processing FFPE samples is inherently challenging due to the template damage incurred during fixation and the presence of residual cross-links. Watchmaker’s novel FFPE treatment step, paired with a reverse transcriptase specifically engineered for RNA-seq applications, delivers excellent sensitivity and enables researchers to address a broader range of clinically relevant samples effectively.
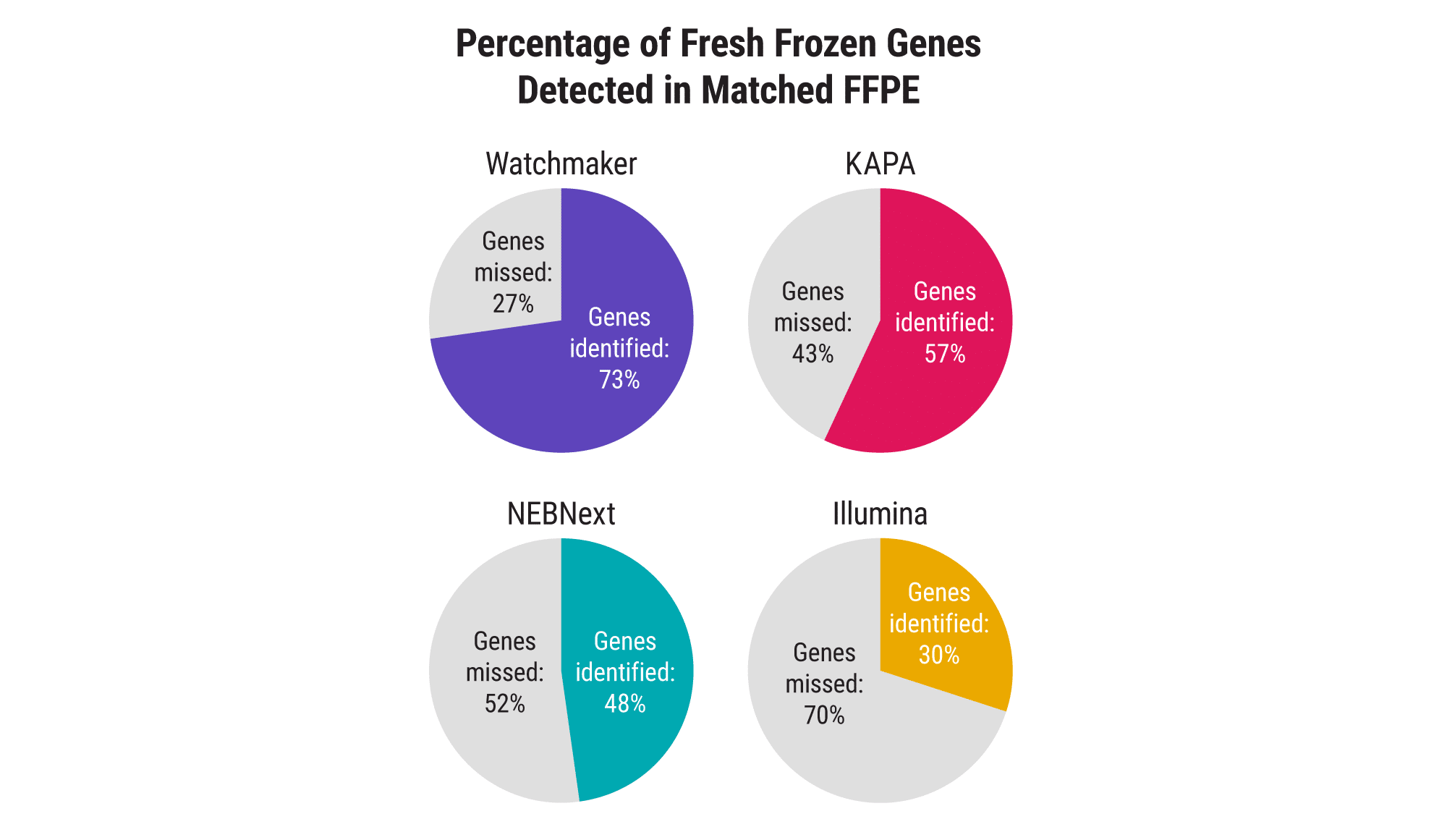
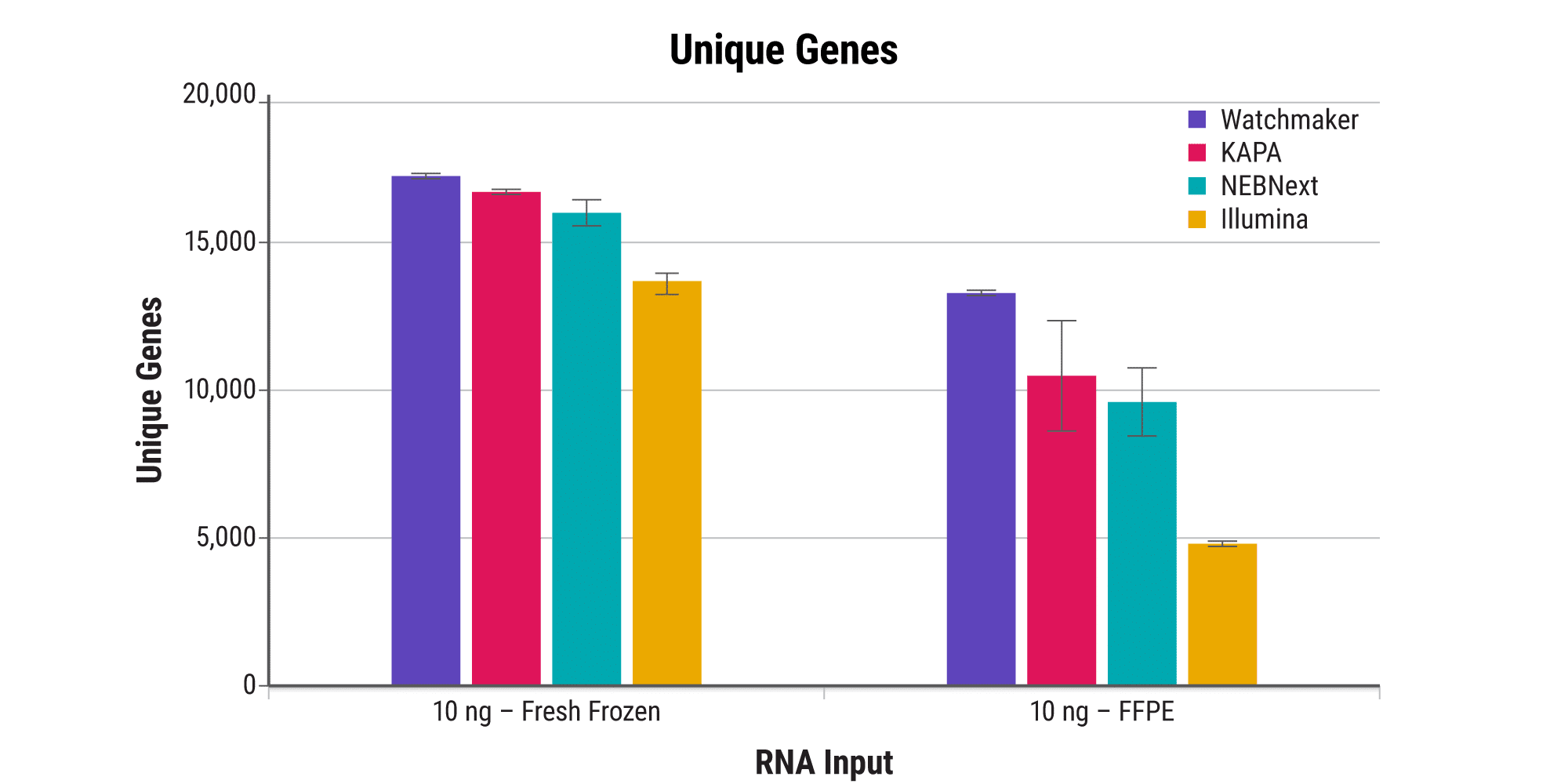
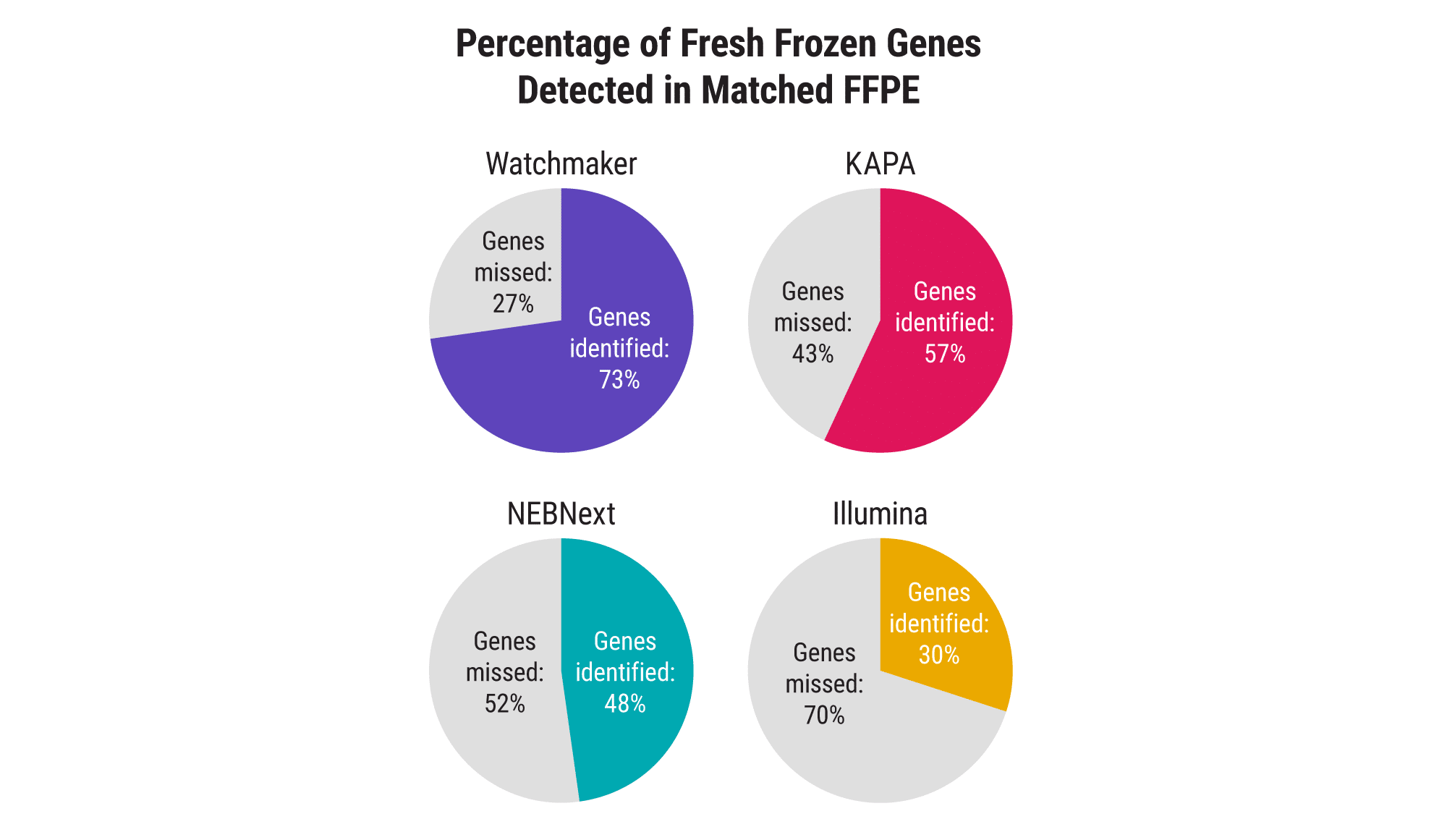

Increase sensitivity with ultra-low-input samples
Robust and reproducible sample processing can be difficult for applications and samples where RNA quantities are restrictive, such as fine needle biopsies, limiting the sensitivity of associated assays. Watchmaker RNA Library Prep Kits with Polaris Depletion enable the generation of high quality libraries with as little as 1 ng of RNA.
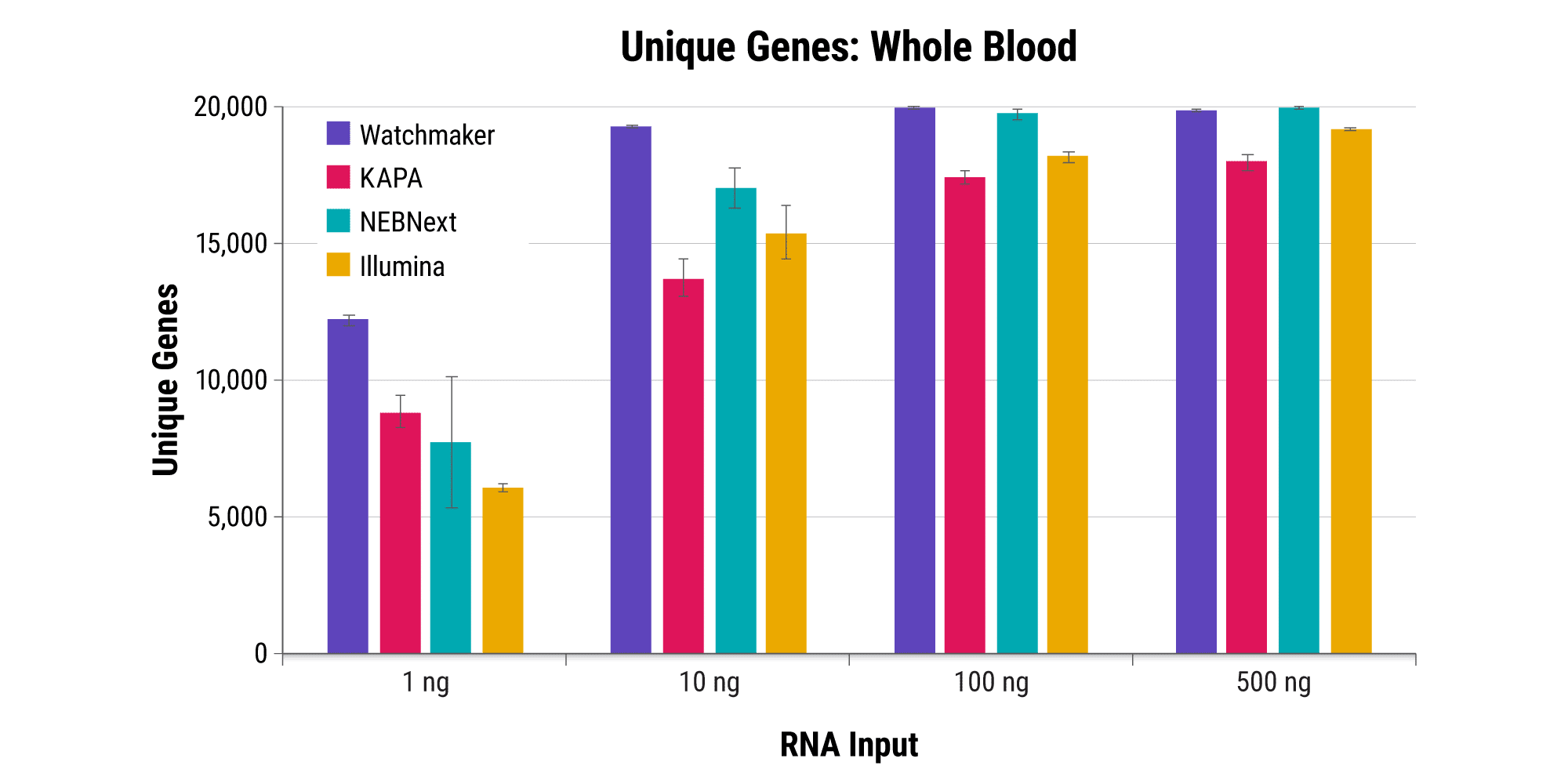
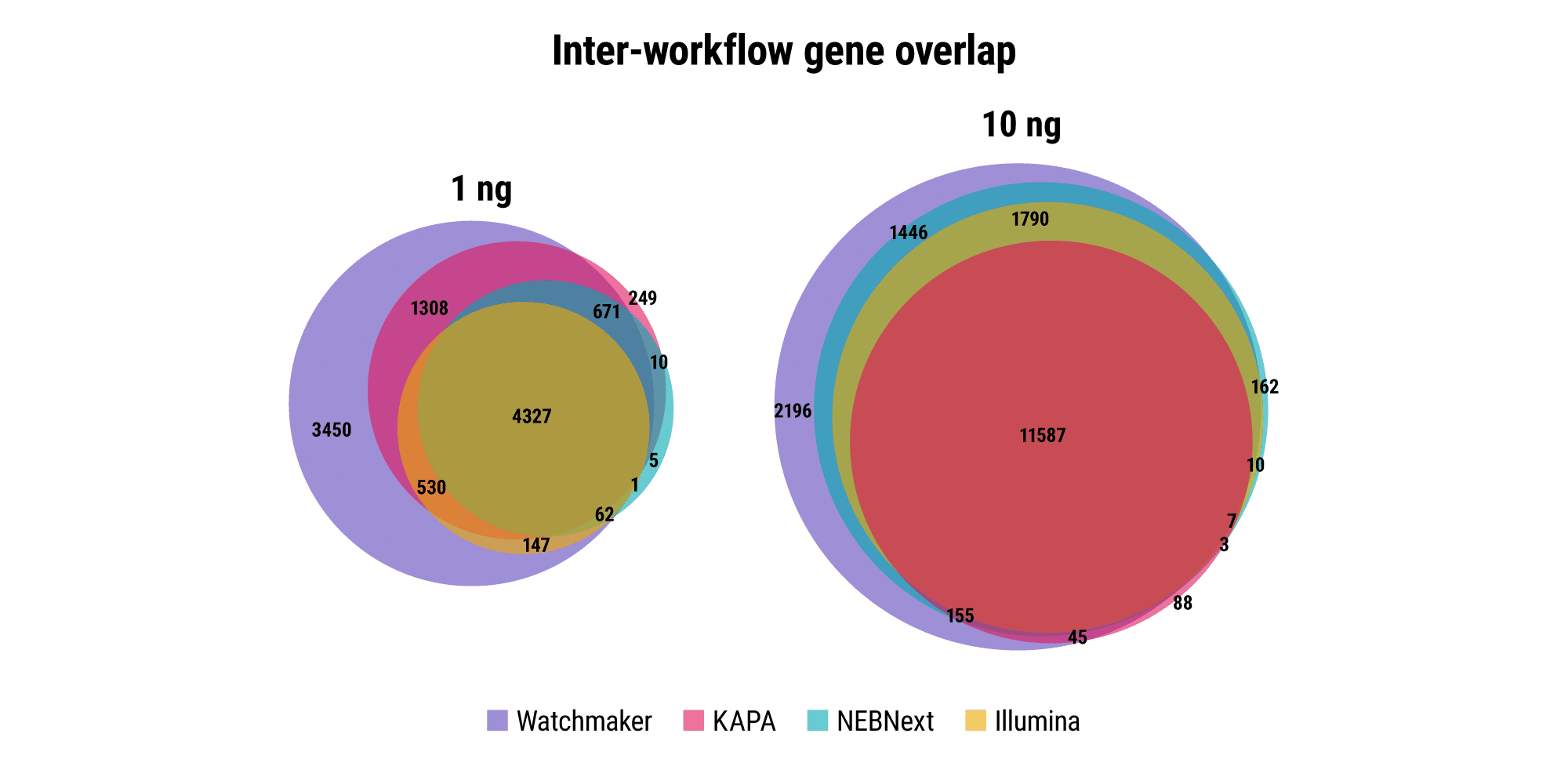



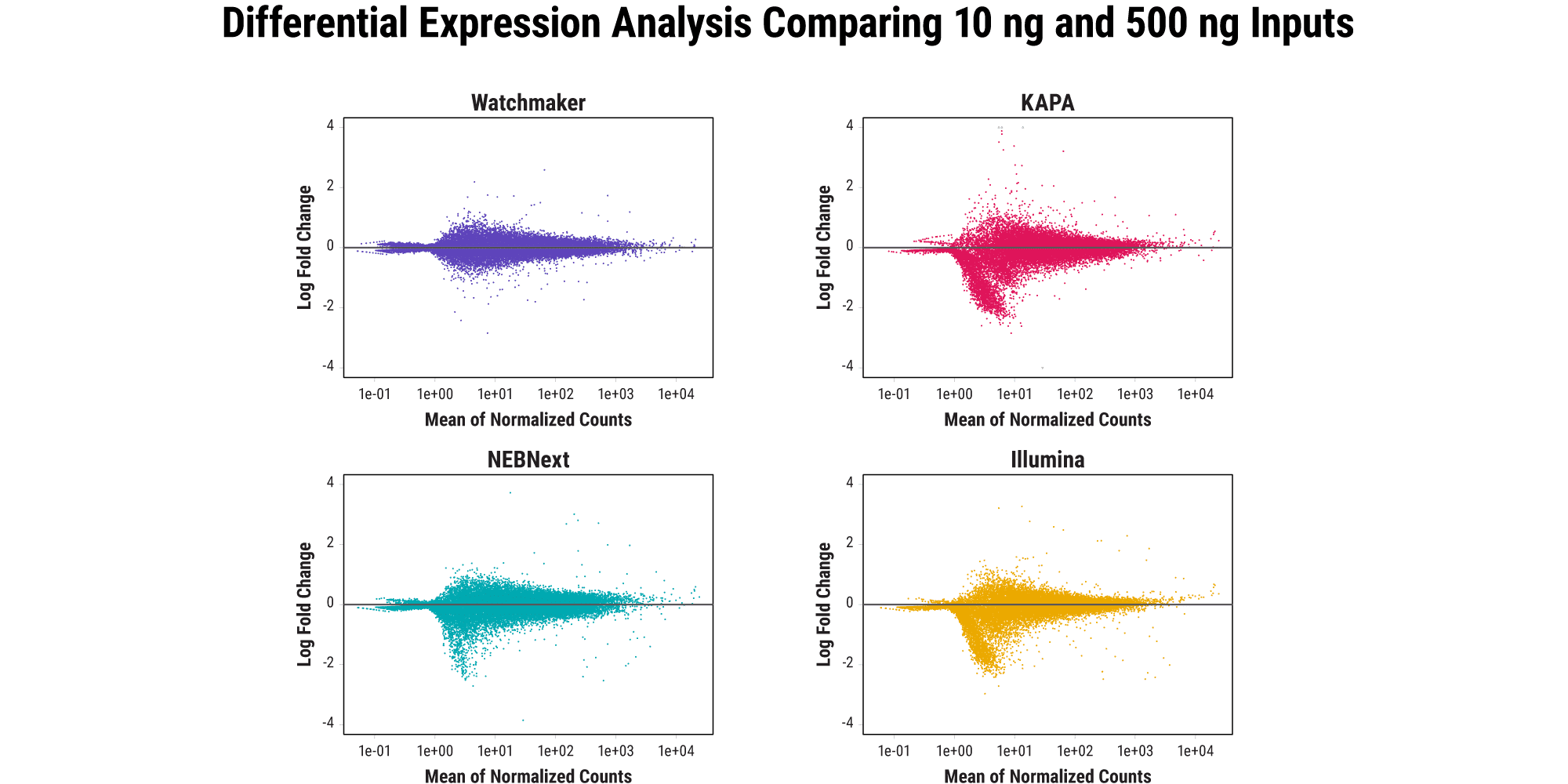
Retain quantitative gene expression information across input amounts
Agreement between high- and low-input libraries with respect to unique genes identified and their abundance is indicative of how well sample complexity is maintained as RNA input amounts decrease. Ideally, the relative gene expression profile should remain consistent across input amounts for a given sample. Watchmaker better preserves the gene expression profile as RNA input amount decreases from 500 ng to 10 ng, whereas other commercial solutions result in data distortion with low inputs.

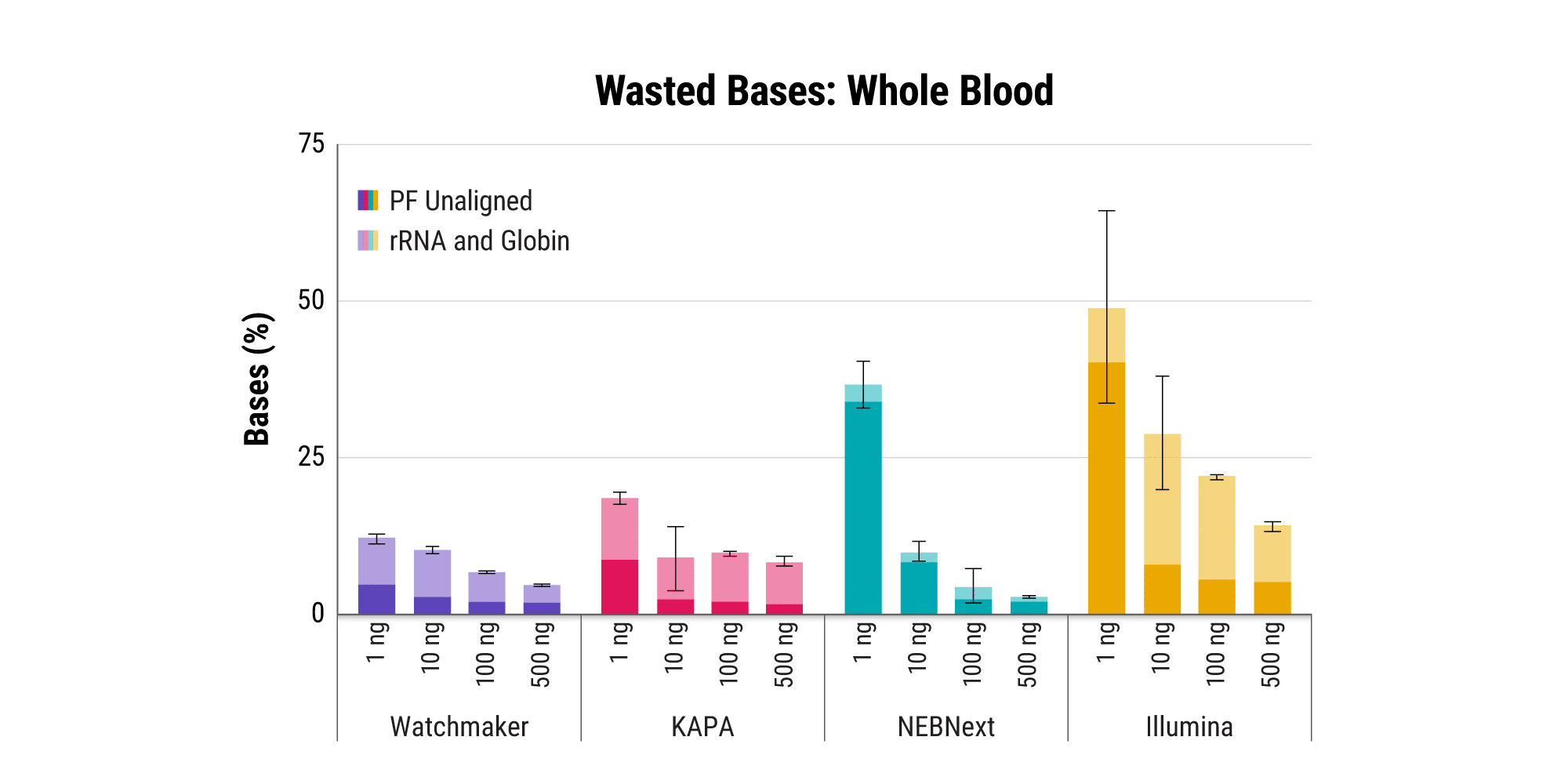
Efficiently sequence blood samples while covering lncRNAs
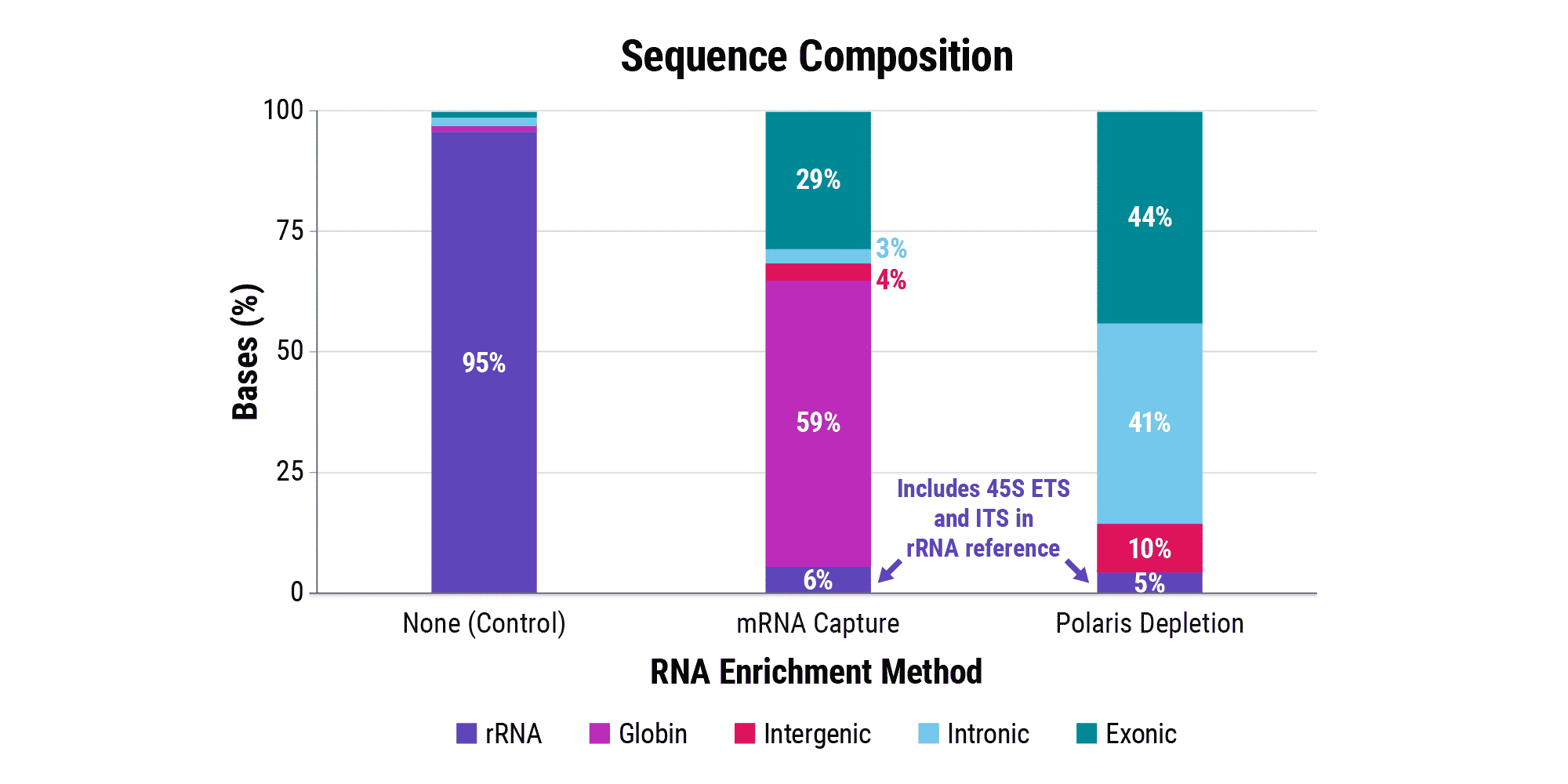
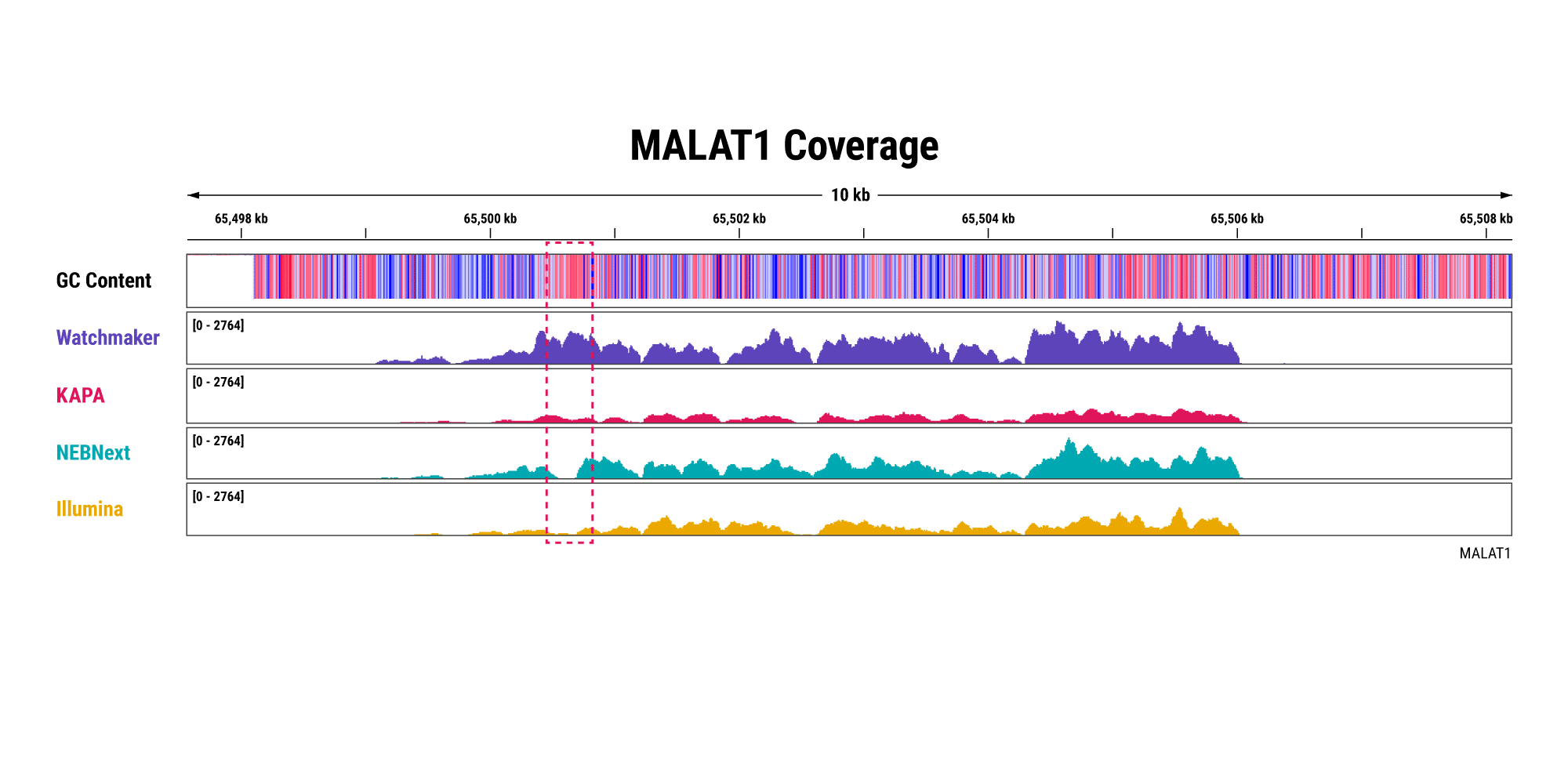
Overabundant globin mRNAs in blood samples pose a challenge for efficient RNA sequencing, as ≥ 30% of bases map to these transcripts if traditional mRNA capture is used. Additionally, mRNA capture omits a significant subset of long non-coding RNAs (lncRNAs) that have demonstrated regulatory functions.¹ Polaris Depletion removes both rRNA and globin mRNA while also providing excellent coverage of lncRNAs, such as MALAT1 which has been implicated in at least 17 cancer types, including lung, breast, and pancreatic.²
- Statello, L., Guo, CJ., Chen, LL. et al. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22, 96–118 (2021).
- Amodio, N., Raimondi, L., Juli, G. et al. MALAT1: a druggable long non-coding RNA for targeted anti-cancer approaches. J Hematol Oncol 11, 63 (2018).