Introduction to whole genome sequencing
Whole genome sequencing (WGS) refers to sequencing the entire genetic blueprint of organisms and viruses and has a variety of applications:
- De novo sequencing to establish new reference genomes
- Low pass WGS as a powerful alternative to microarrays for NGS-based genotyping
- Human resequencing to elucidate the genetic basis of inherited, rare, complex, metabolic, and pediatric diseases – especially those associated with large and/or previously uncharacterized structural variation (e.g., deletions, insertions, inversions, duplications, translocations, and copy number variation)
Sonication has long been regarded as the gold standard for unbiased DNA fragmentation and data. However, it can be a bottleneck in high throughput labs and comes with expensive equipment and consumable costs. Enzymatic DNA fragmentation is an attractive alternative that scales easily and minimizes sample loss.
Key considerations when selecting a WGS workflow
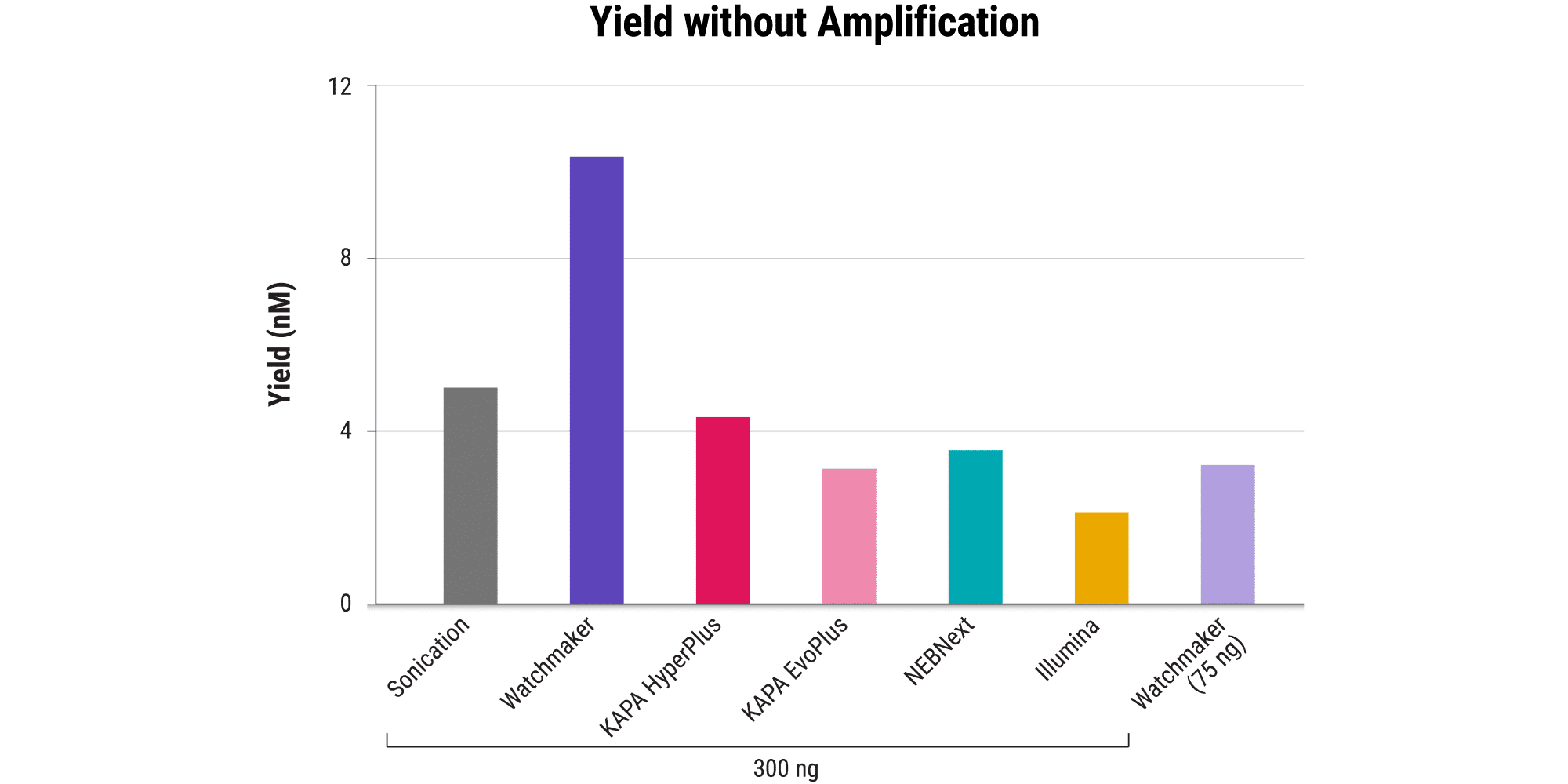
Minimizing bias: Compatibility with PCR-free sequencing
- Library amplification can be a source of both fragment length and sequence bias – where shorter fragments and GC-neutral sequences are preferentially amplified.
- When performing PCR-free sequencing, the core library prep workflow needs to be highly efficient to generate sufficient yields for sequencing without amplification.
- The Watchmaker Library Prep Kit with Fragmentation generates the highest PCR-free yields, enabling the use of lower DNA inputs and broadening the addressable sample range for researchers.

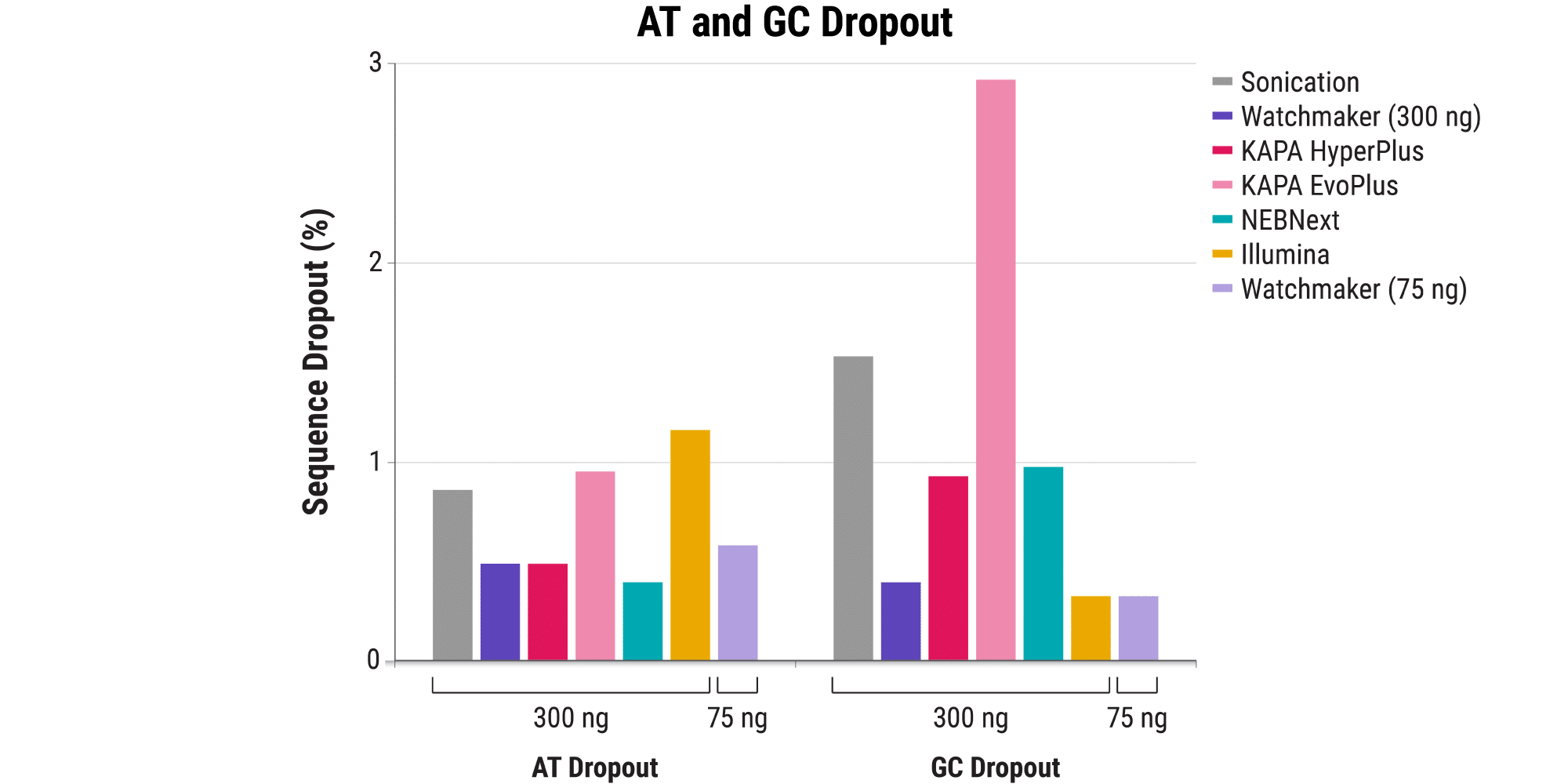
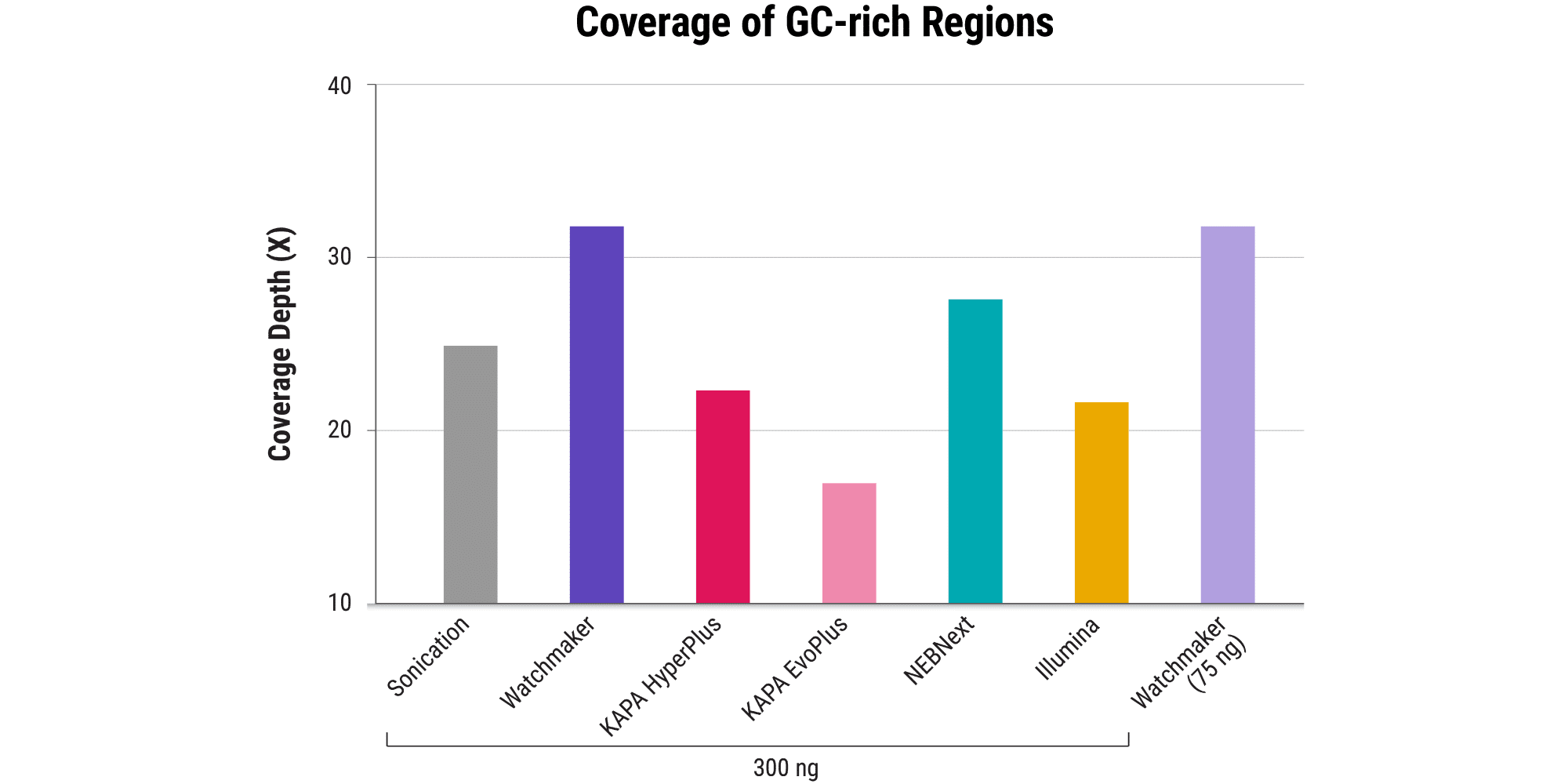
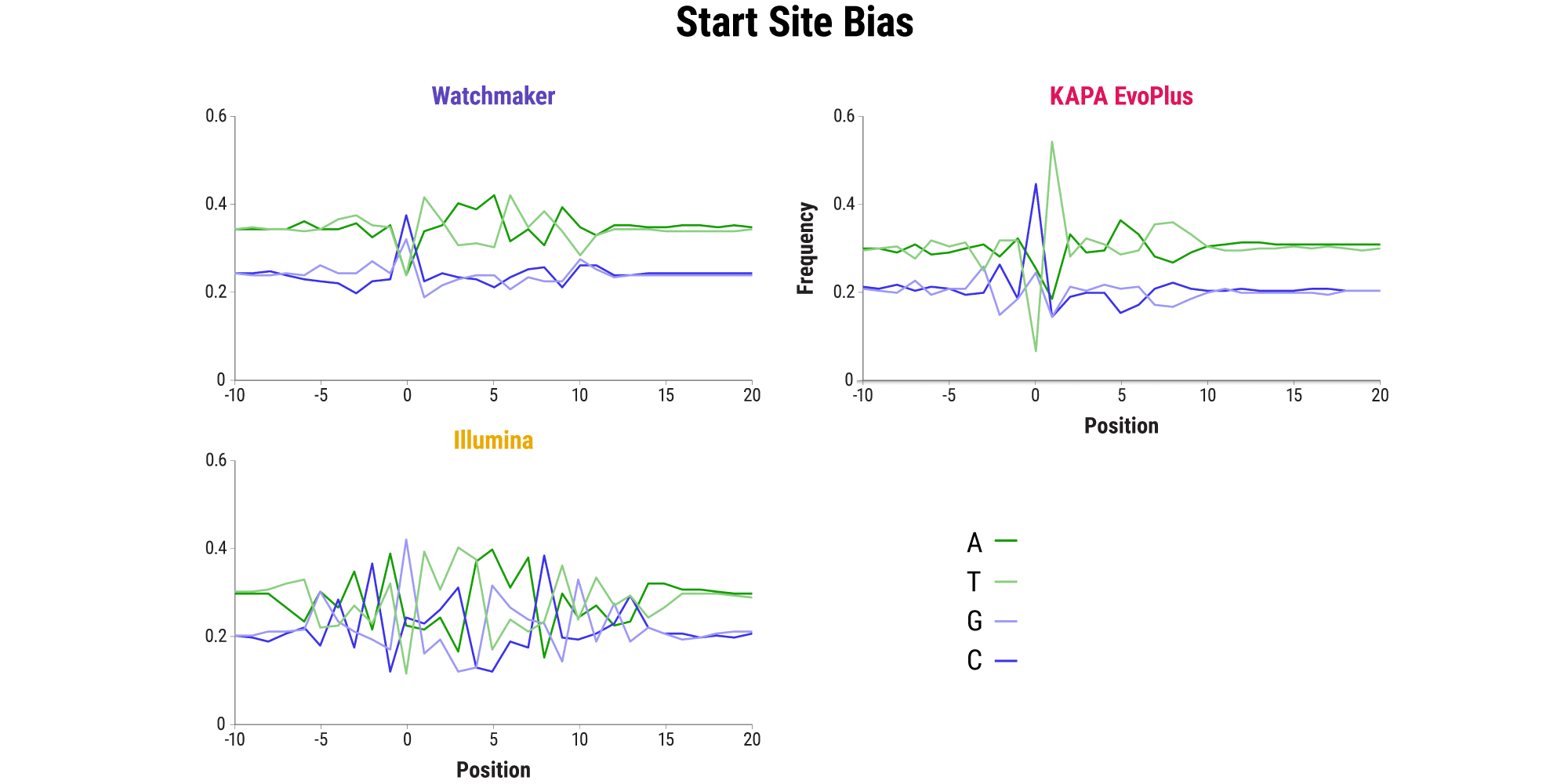
Minimizing bias: Maintain a clear picture of the whole genome
- Even in the absence of PCR, sequence bias, including start site bias, can lead to uneven genome coverage and result in certain regions being underrepresented or completely missed.
- The Watchmaker Library Prep Kit with Fragmentation delivers minimal start site bias.
- This results in less AT/GC dropout and improved coverage of challenging GC-rich promoters in comparison to other solutions – including a sonication control.



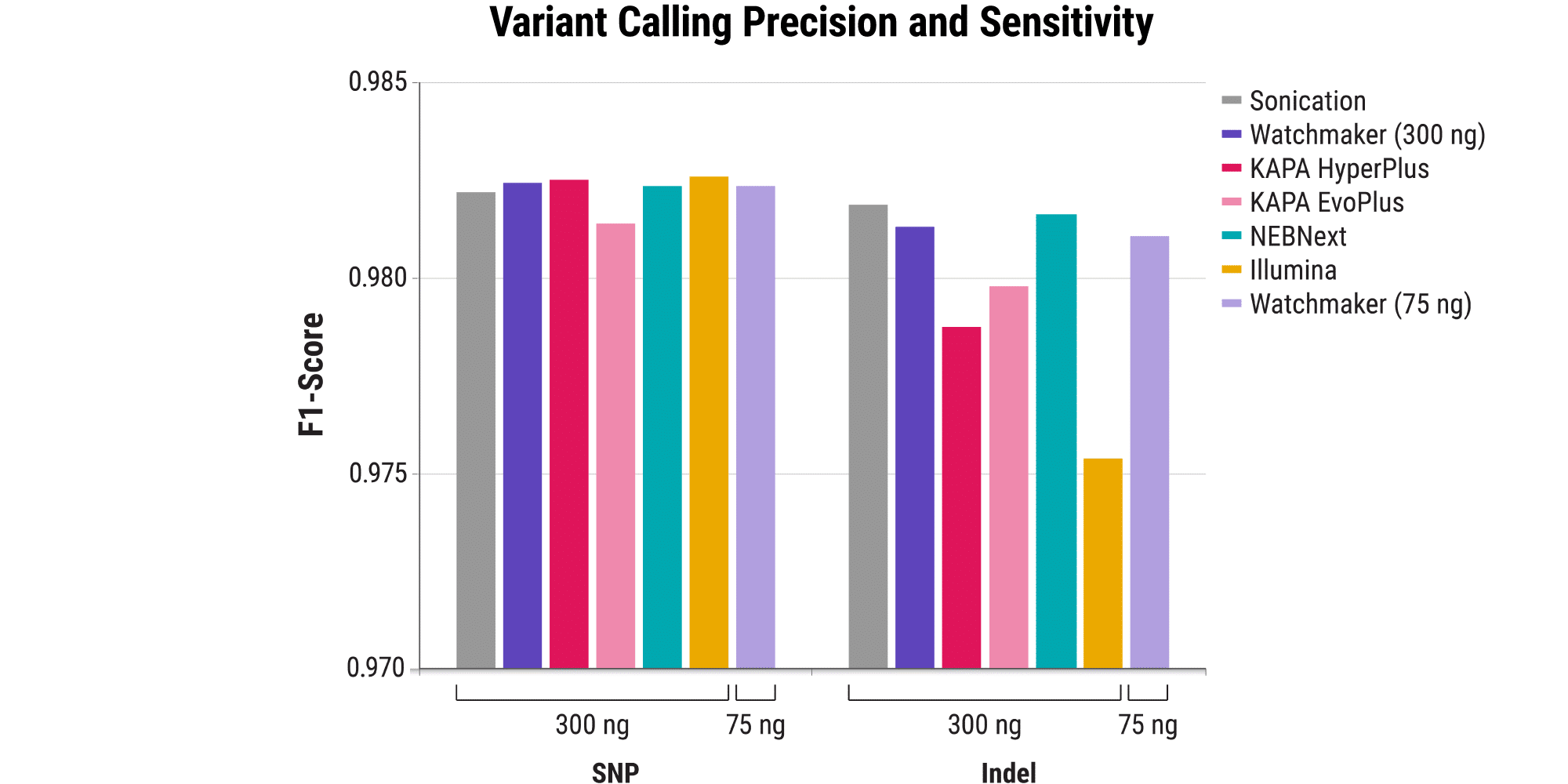
Maintaining data quality: Robust variant calling
- Many genetic diseases are caused by specific mutations in the genome.
- Sensitive and precise variant calling is essential for identifying disease-causing mutations.
- The Watchmaker DNA Library Prep Kit with Fragmentation delivers excellent F1 scores for SNP and indel variant calling (a combined measure of precision and sensitivity) on-par with sonication.
- Available resource: AGBT 2022 poster (Broad Institute)

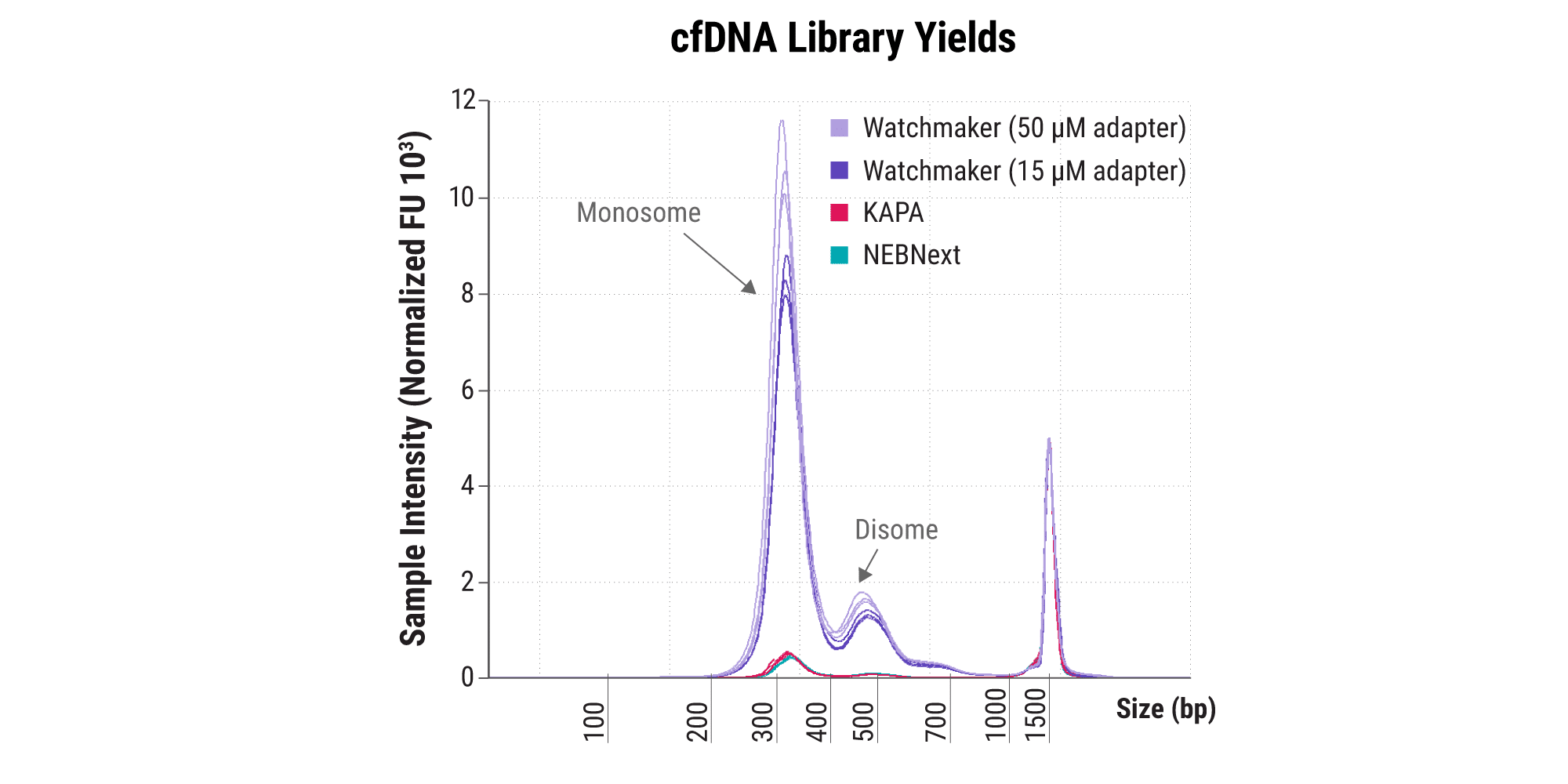
Compatibility with clinically relevant sample types
- WGS of cancer genomes from cell-free DNA (cfDNA) and FFPE samples can be performed to survey somatic mutations at different stages of disease development.
- The Watchmaker DNA Library Prep Kit improves the conversion of low-input cfDNA to final library – translating to improved library complexity and coverage.
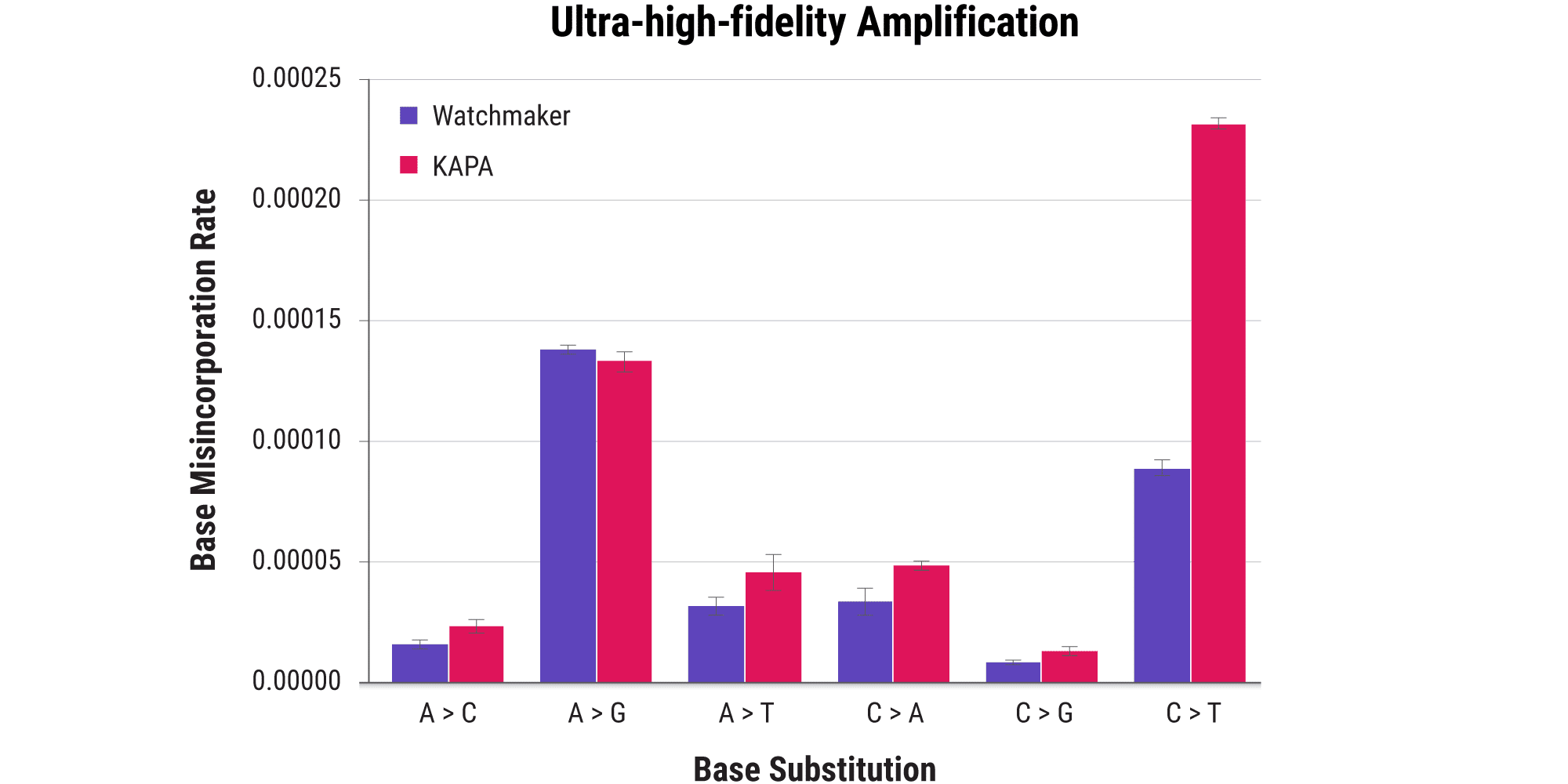
- For samples like cfDNA and FFPE where PCR is required, it’s critical to use a polymerase system that is highly efficient (to minimize cycle counts), low-bias, and high-fidelity (to minimize PCR errors).
- Equinox Amplification Master Mix is designed to deliver in all of these areas, limiting bias and significantly reducing base misincorporation events.


For assay developers – make realizing your next product easier
- Navigate the complexities of productization with an experienced team who’s as committed to your success as you are
- All aspects of our customization process are designed to serve you with speed, agility, and above all else, a commitment to quality
- Tailored fill volumes, labeling (including white and private label), and packaging designed to your specifications
- All products are manufactured within an ISO 13485:2016-certified QMS (download our certificate)
Discover more with Watchmaker
Watchmaker DNA Library Prep Kits with Fragmentation
A streamlined single-tube chemistry with reduced bias and artifacts. Ideally suited for PCR-free workflows, ultra-low inputs, and low-quality DNA samples including FFPE
Watchmaker DNA Library Prep Kits
A simple workflow that improves yields and coverage of low-input DNA samples, such as cfDNA.
Equinox Library Amplification Kits
An ultra-high-fidelity DNA polymerase in an optimized hot start master mix format, specifically optimized for high-efficiency, low-bias NGS library amplification