What are the recommended applications for the Watchmaker DNA Library Prep Kit with Fragmentation?
The kit is designed for highly efficient conversion of variable quality DNA into Illumina-compatible sequencing-ready libraries so it supports a variety of applications including:
- Whole genome sequencing (WGS)
- Whole exome sequencing (WES)
- Targeted sequencing using hybridization capture
- Low-input and challenging samples such as FFPE
- Metagenomic analysis
- RNA sequencing (using cDNA as input)
Should I really vortex the Enzymatic Fragmentation, End Repair and A-tailing (Frag/AT) reactions?
Yes. Vortexing the master mix components, master mix, and Frag/AT reaction ensures consistent fragmentation results between samples. When preparing a small number of reactions, it is acceptable to mix by pipetting.
Is the Watchmaker DNA Library Prep Kit with Fragmentation compatible with FFPE DNA?
The kit is compatible with DNA extracted from FFPE. DNA extracted from FFPE may have variable degrees of crosslinking, or other chemical modifications, which may affect fragment length and/or conversion efficiency of input DNA into sequencing library. Ensure the extraction protocol is optimized for the sample type. Assessing DNA quality by qPCR prior to beginning library preparation can inform input mass requirements for the desired application.
Shorter fragmentation times may be required for FFPE to meet the same fragment length profiles achieved with intact DNA. As a starting point, decrease the fragmentation time by half to achieve the same insert size as for intact DNA. Further optimization may be required.
How should I modify enzymatic fragmentation conditions as I increase/decrease the amount of input DNA?
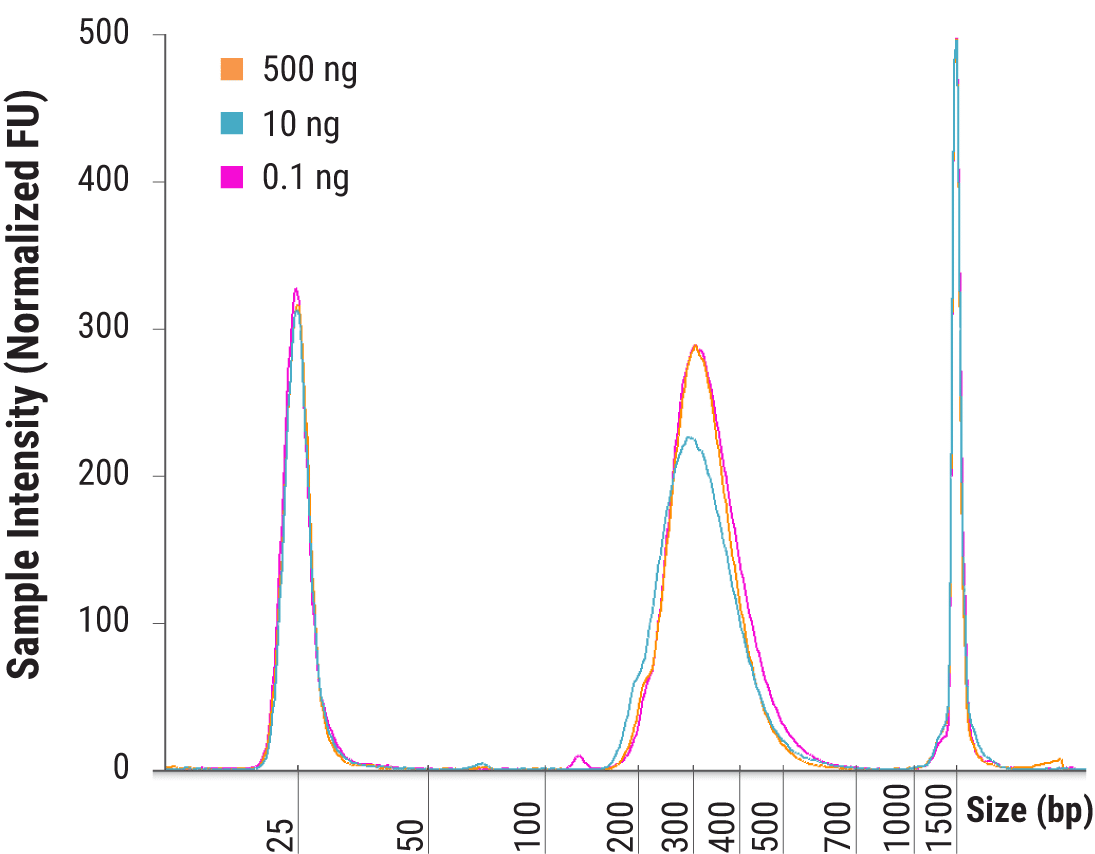
Libraries were constructed in duplicate from 500, 10, and 0.1 ng of human genomic DNA fragmented for 20 minutes at 37°C. Final library size distributions were assessed using the D1000 assay by TapeStation (Agilent).
How should I adjust the adapter concentration for different input amounts?
| DNA input (ng) | Adapter concentration* | Adapter:insert molar ratio |
|---|---|---|
| 10 – 500 | 15 µM | Up to 1500:1 |
| 1 – 10 | 3 µM | 300 – 3000:1 |
| < 1 | 0.6 µM | > 300:1 |
*Adapter to insert ratio estimated based on 300 bp fragment sizes; input amounts over 50 ng will use the same volume and concentration of adapter; for masses under 10 ng, calculate the concentration required to maintain an adapter to insert concentration of 300:1.
What is the recommended input mass for the kit?
The kit is compatible with a broad range of input amounts (1 ng – 500 ng). Conversion efficiencies should be high across this range when working with high quality DNA inputs.
How much input do I need to generate sequenceable concentration of library with a PCR-free workflow?
The optimal input range will depend on the application. With an estimated target library concentration of 5 nM, we suggest using at least 20 ng – 30 ng as a starting point when generating libraries with 200 bp insert lengths (e.g., WES). For WGS workflows targeting >400 bp inserts lengths, we recommend starting with ~100 ng of input DNA. Yields may vary depending on the quality of the DNA going in (FFPE requires more mass input).
Any special considerations for PCR-free?
Ensure enough DNA input is used to create enough library to QC and sequence, without amplification. 100 ng of high quality DNA is a good place to start, although it is possible to generate high quality libraries from 50 ng or less. FFPE DNA may require more input, and will vary from sample to sample. Full length adapters are required, as there is no opportunity to introduce sample barcodes or P5/P7 sequences into the library via PCR.
Note: PCR-free libraries appear to be longer than they truly are on electrophoresis systems like TapeStation and Bioanalyzer due to the structure of the adapter.
What adapters are compatible with the Watchmaker DNA Library Prep Kit with Fragmentation?
Any adapter with a 3′ overhang T is compatible. Note that adapter quality impacts overall library preparation efficiency.
Here are some options from IDT:
- xGen Stubby Adapter and UDI Primers (Catalog number 10005976 or 10005921). This solution involves ligating a stubby or truncated Y adapter to the library during ligation, then adding in unique dual indexes with PCR primers during library amplification to incorporate sample barcodes for multiplexed sequencing.
- xGen UDI-UMI Adapters (Catalog number 10006914 and 10005903). These are full-length Y-adapters (unique dual sample barcodes, with optional UMIs) which are ligated. These can be used for PCR-free applications if sufficient input is used in library construction.
For the fragmentation workflow we currently run, all samples go through a 3X SPRI bead cleanup before beginning fragmentation to ensure samples are all eluted in Tris and not TE. Do you recommend keeping this bead clean up in our protocol for the best fragmentation results?
EDTA can affect fragmentation, depending on final concentration in the reaction. It is tolerated to 0.1mM in the final 50ul reaction. If EDTA would exceeds 0.1mM in 50 µL, a cleanup is recommended.
Can I use PCR products or amplicons with the Watchmaker DNA Library Prep Kit with Fragmentation?
Yes, the fragmentation times recommended in the user guide are applicable for gDNA as well as PCR products so no further optimization is required. Fragmentation of high-quality human gDNA, plasmid DNA (~5 kb), and a 1.1 kb PCR product in the same experiment, with the same fragmentation conditions, resulted in comparable fragment lengths.
My fragment sizes are smaller than expected. How should I handle the sample so that my fragment lengths match what is expected in the User Guide?
There are two things to focus on to help achieve fragment lengths in-line with the recommendations in the User Guide. The goal is to ensure the reaction does not begin until it is placed into the thermal cycler. This is accomplished by keeping all components of the reaction ice-cold during preparation. Some pointers to facilitate:
- Equilibrate the fragmentation enzyme and buffer on ice.
- Ensure the DNA sample (in 40 µL) is pre-chilled on ice. Pay special care to allow the sample to cool if the diluent used to dilute the stock DNA was stored at room temperature. We find these to be very effective.
- Add the chilled enzymes and buffer to the DNA sample on ice.
- When mixing the reaction prior to thermal cycling, perform this step quickly to minimize any potential elevation of temperature. Keep the reaction on ice until transferring directly to the pre-chilled thermal cycler. Ensure ramp rates on the thermal cycler are set as default (as opposed to a slower ramp rate).
What is typical user to user, lot to lot, day to day variability for fragment lengths?
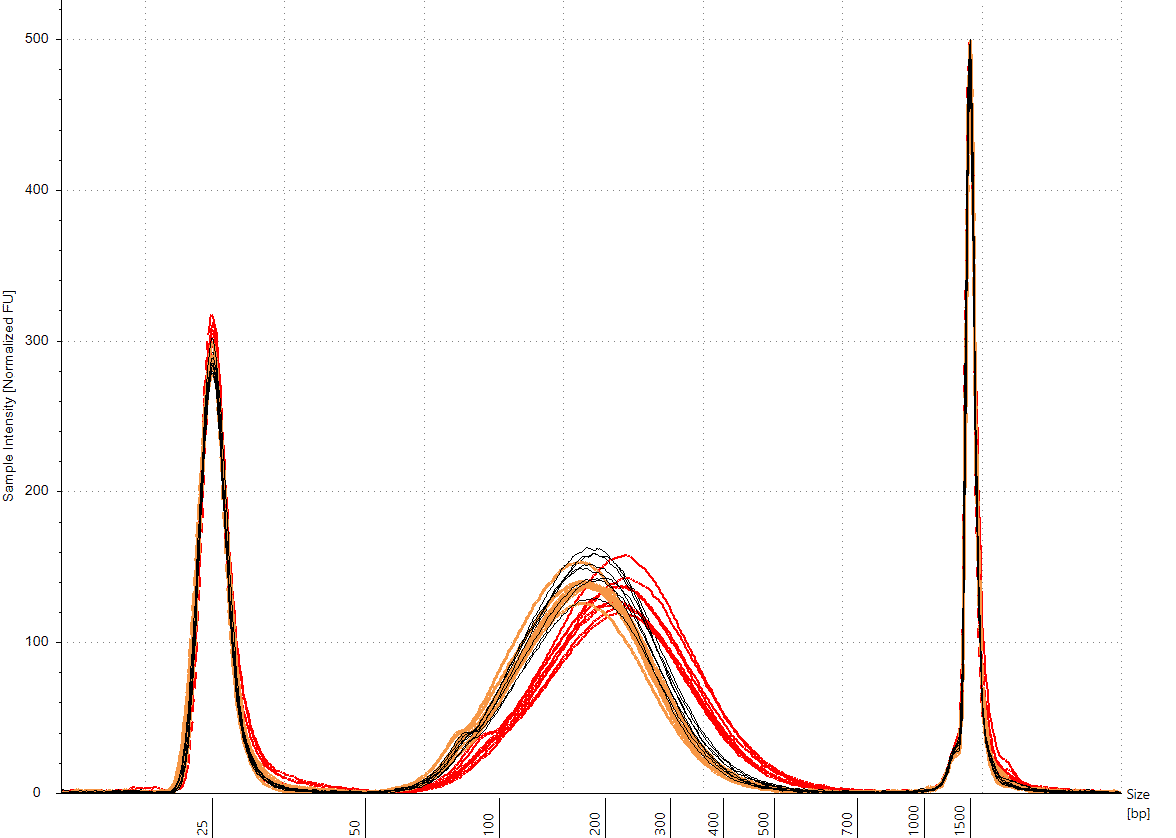
- Technical replicate CVs are less than 3%
- The deviation from the target of 200 bp is 6 – 8%
- User-to-user and day-to-day variance is less than 4 bp
- Lot-to-lot variance was about 25 – 30 bp (in both cases less than 8% from the target)
Do you offer custom formats?
Yes! Watchmaker offers customer fills, packaging, concentrations, and labeling – including private label – designed to meet your unique needs. We offer flexible terms to serve organizations of any size, and our right-sized processes enable rapid turnaround time on customization. Please contact sales@watchmakergenomics.com to learn more about our capabilities.
Are you ISO 13485-certified?
Yes! Our Quality Management System has achieved ISO 13485:2016 Certification. Our certificate has been awarded for the design, development, manufacture, contract manufacture, and support of high performing reagents for genomics applications in medical research. Download the certificate here.